
Cell | Pan-cancer single-cell dissection reveals phenotypically distinct B cell subtypes
On July 23rd, 2024, the Zhang Lab from the Biomedical Pioneering Innovation Center (BIOPIC) at Peking University published a paper entitled “Pan-cancer single-cell dissection reveals phenotypically distinct B cell subtypes” in Cell. Analyses of tumor-infiltrating B cells (TIBs) from patients across 19 cancer types depict their abundance and subtype heterogeneity at the pan-cancer level and identify tumor-associated atypical B cells(TAABs)that can communicate with CD4 T cells and have prognostic potential.

The success of cancer immunotherapy has inspired the in-depth exploration of the holistic tumor ecosystem, especially the immune aspect of the tumor microenvironment.1 The comprehensive chart of the functional states and heterogeneity of tumor-infiltrating T, myeloid and NK cells2–4 has enlightened multiple novel therapeutic strategies.However, despite their important impact on immunity,5 B cells have received disproportionately less attention.
This studyestablished a comprehensive resource of human B cells by integrating single-cell RNA-sequencing data of511,847cells from 649 patients across 19 major cancer types. Five B cell major clusters and 20 fine-grained B cell subtypes were identified through batch correction and unsupervised clustering.The researchers additionally computationally reconstructed the BCR sequencesof individual B cells with available raw sequencing dataand conducted TIB-related immunohistochemistry staining on 80 specimens from patients diagnosed with one of nine cancer types.Such a resource enabled the systematic inspection of the transcriptional, clonotypic and spatial heterogeneity of TIBs across cancer types.
Based on the integrative data, the abundanceand subtype composition of TIBs exhibited substantial heterogeneity acrossdifferent cancer types.This atlas also enabled the identification of critical transitional subsets amidst B cell differentiation stages, such as pre-germinal centerB cells and plasmablasts, within tumors.Particularly, the researchers demonstrated the occurrence of germinal center reactions within tumors akin to secondary lymphoid organs, with the process of the iterative BCR sequence evolution and the genesis of both memory B cells and antibody-secreting cells recapitulated. In addition, a skewness towards IgG, especially IgG1, in the isotypes of intratumoral antibody-secreting cells was demonstrated.
Moreover, the researchers identified stress-response memory B cells and TAABs, two pan-cancer-shared tumor-enriched subpopulations associated with shortened and prolonged survival of cancer patients at the pan-cancer level respectively.The signature of stress-response memory B cell subsetresembled the previously reported tumor-associated dysfunctional NK and stress-response T cells, whose stress-response signatures have also been linked with poor survival or immunotherapy resistance.4,6Interestingly, in the attempt to pinpoint TIB subsets associated with unfavorable patient outcomes, the researchers found that regulatory B cells, or at least IL10+ B cells, remained elusive in the single-cell clustering analysis and potentially arose from various stages of B cell differentiation.
In particular,the tumor-enriched TAABs featured high clonal expansion levels and proliferative capacity as well as a highly activated transcriptional state within tumors. CD4 T cells, especially tumor-reactive CXCL13+ CD4 T cells, might cooperate with TAABs in their activation and differentiation toward plasma cells.Importantly, the TAAB signature was linked with patient prognosis in a cancer type-dependent way, and alsoserved as a predictor of favorable response tomultiple immunotherapy strategies.
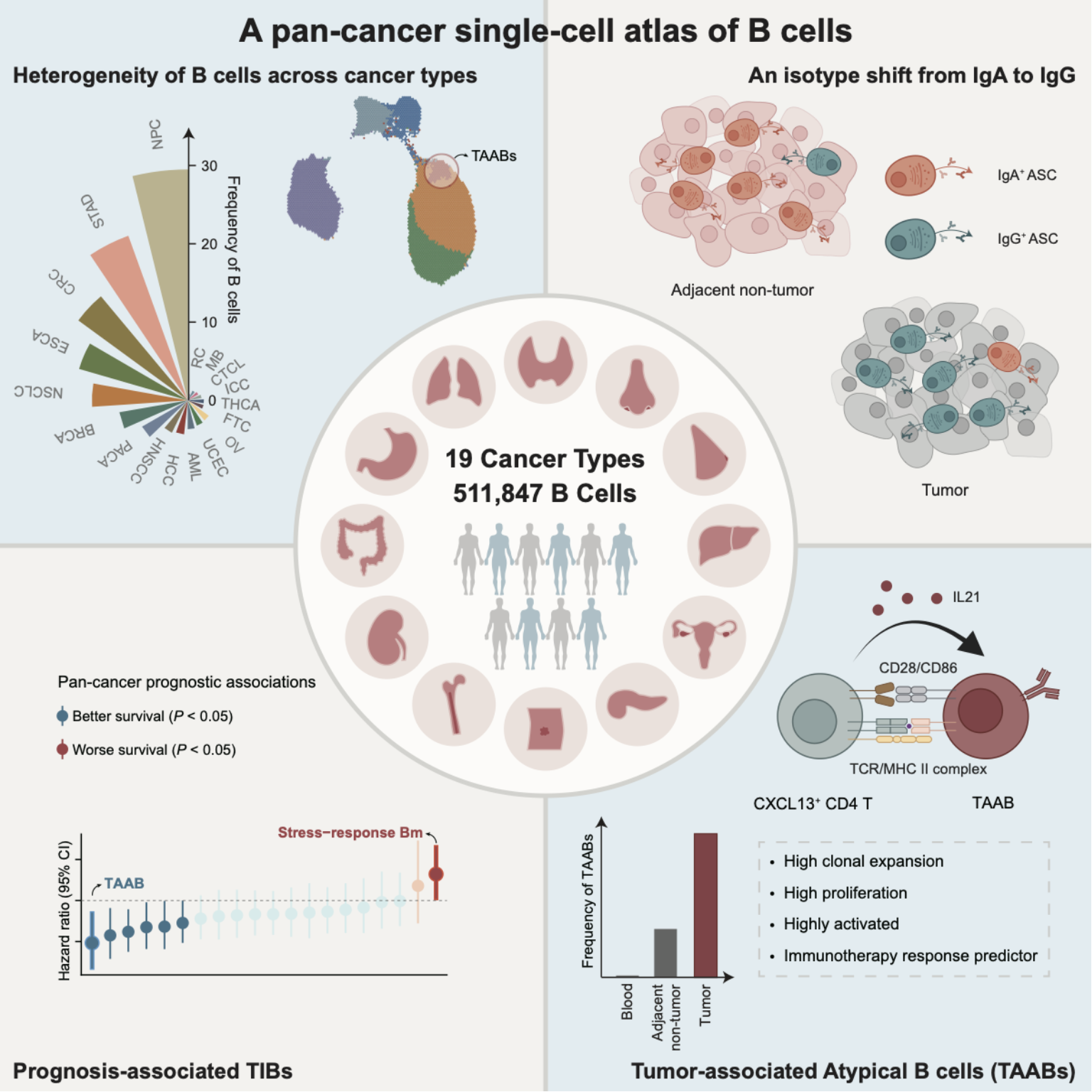
Figure 1. The graphical abstract of thisstudy.
In summary, this integrative resource improves the current understanding of B cells from a pan-cancer perspective, shedding new light on the heterogeneity of B cells and their anti-tumor immune response andlaying a foundation for further exploration of functional commonality and diversity of B cells in cancer.
Ph.D. candidate Yu Yang and Ph.D. candidate Xueyan Chen are the co-first authors of the paper. Prof. Zemin Zhang, Dr. Min-Min Chen and Dr. Dongfang Wang are the co-corresponding authors of the paper. The project was supported by funding from the National Natural Science Foundation of China, Beijing Municipal Science and Technology Commission, National Key Research and Development Program of China, Major Program of Shenzhen Bay Laboratory, and Open Program of Shenzhen Bay Laboratory.
Link: https://www.cell.com/cell/fulltext/S0092-8674(24)00712-8
Reference:
1.Wang, D.F., Liu, B.L., and Zhang, Z.M. (2023). Accelerating the understanding of cancer biology through the lens of genomics. Cell 186, 1755–1771. https://doi.org/10.1016/j.cell.2023.02.015.
2.Zheng, L., Qin, S., Si, W., Wang, A., Xing, B., Gao, R., Ren, X., Wang, L., Wu, X., Zhang, J., et al. (2021). Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 374, abe6474. https://doi.org/ARTN eabe6474 10.1126/science.abe6474.
3.Cheng, S., Li, Z., Gao, R., Xing, B., Gao, Y., Yang, Y., Qin, S., Zhang, L., Ouyang, H., Du, P., et al. (2021). A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell 184, 792-809.e23. https://doi.org/10.1016/j.cell.2021.01.010.
4.Tang, F., Li, J., Qi, L., Liu, D., Bo, Y., Qin, S., Miao, Y., Yu, K., Hou, W., Li, J., et al. (2023). A pan-cancer single-cell panorama of human natural killer cells. Cell 186, 4235-4251.e20. https://doi.org/10.1016/j.cell.2023.07.034.
5.Morgan, D., and Tergaonkar, V. (2022). Unraveling B cell trajectories at single cell resolution. Trends Immunol. 43, 210–229. https://doi.org/10.1016/j.it.2022.01.003.
6.Chu, Y., Dai, E., Li, Y., Han, G., Pei, G., Ingram, D.R., Thakkar, K., Qin, J.-J., Dang, M., Le, X., et al. (2023). Pan-cancer T cell atlas links a cellular stress response state to immunotherapy resistance. Nat. Med. 29, 1550–1562. https://doi.org/10.1038/s41591-023-02371-y.