
Genome Biology | Parallel single-cell and bulk transcriptome analyses reveal key features of the gastric tumor microenvironment
The tumor microenvironment (TME) has been shown to strongly influence treatment outcome for cancer patients in various indications and to influence the overall survival. However, the cells forming the TME in gastric cancer have not been extensively characterized.
On December 22nd, 2022, the Zhang Lab from Biomedical Pioneering Innovation Center (BIOPIC), Beijing Advanced Innovation Center for Genomics (ICG), School of Life Sciences in Peking University published a paper titled “Parallel single-cell and bulk transcriptome analyses reveal key features of the gastric tumor microenvironment” in Genome Biology. The published study combined bulk and single-cell RNA sequencing from tumors and matched normal tissue of 24 treatment-naïve gastric cancer patients and elucidated the characteristics of 81 cell subtypes in gastric cancer microenvironment. The study finds that activated fibroblasts and endothelial cells are most prominently overrepresented in tumors. Intercellular network reconstruction and survival analysis of an independent cohort imply the importance of these cell types together with immunosuppressive myeloid cell subsets and regulatory T cells in establishing an immunosuppressive microenvironment that correlates with worsened prognosis and lack of response in anti-PD1-treated patients. In contrast, a subset of IFNγ activated T cells and HLA-II expressing macrophages are linked to treatment response and increased overall survival. The published study provides a valuable resource for characterization of gastric cancer microenvironment and identification of new patient stratification biomarkers, and insights for gastric cancer biology and treatment research.
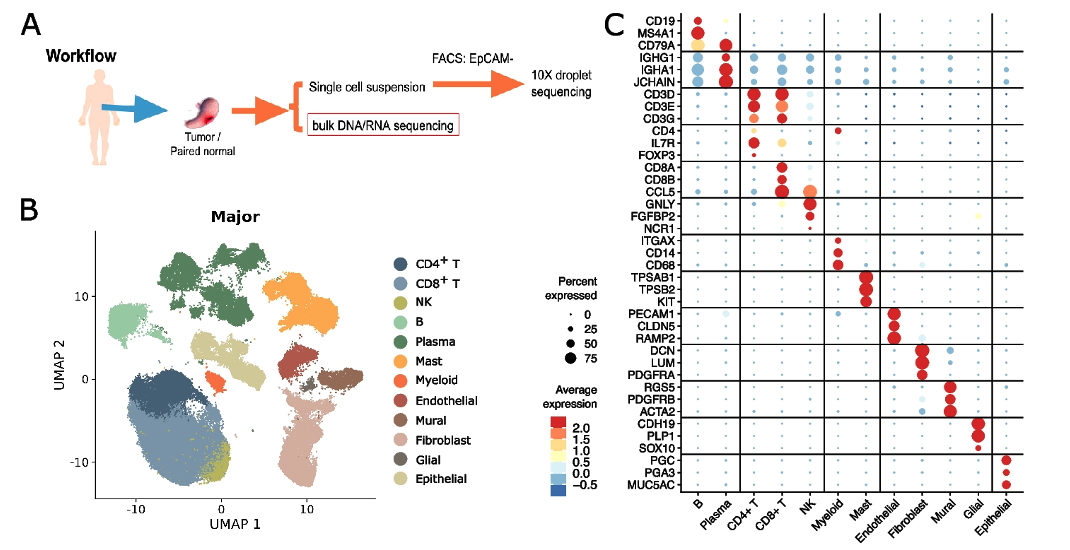
Figure 1 Cellular landscape of EPCAM-negative cells in non-malignant and malignant gastric patient samples.
The researchers performed bulk tissue RNA sequencing (RNA-seq) and whole exome-sequencing together with single-cell RNA-seq (scRNA-seq) of tumor and matched non-malignant gastric tissue samples from 24 treatment-naive gastric cancer patients and identified eleven major cell types and 81 subtypes. Through parallel single cell and bulk RNA-seq analysis, the researchers quantified the change in abundance and cellular activity associated with malignant transformation for each of the stromal cell types. A few of the stromal cell types display strong association with malignant transformation and exhibit unique transcriptional profiles. The activated fibroblast population F13-CTHRC1 is found to be enriched in tumor and has very strong activated myofibroblast characteristics similar to what’s reported in other publications. Its gene signature also displays association with worsened survival in multiple cancer indications. On the other hand, the activated endothelial cell sub types such as EN10-SERPINE1 and EN03-ESM1 are also enriched in tumor tissue. Their activity of angiogenic and cell motility response pathways are enhanced while antigen-presenting and immune defense processes reduced. The gene signatures of these activated endotheilal cells are also associated with worsened prognosis. The identification of these important cell subtypes reveal an important role of the non-malignant stromal cells in the process of gastric tumor pathogenesis and metastasis, and suggests potential therapeutic targets.
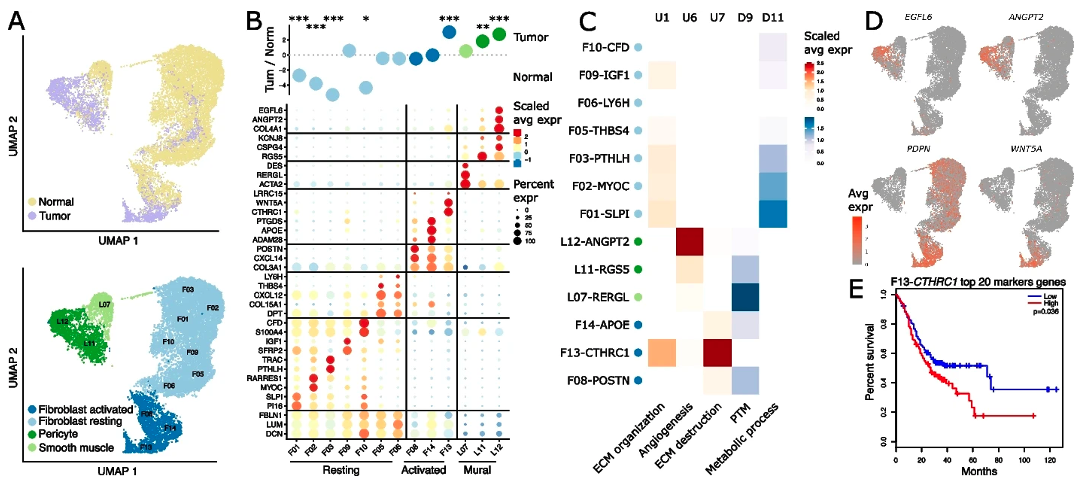
Figure 2 Transcriptional reprogramming in cancer-associated fibroblasts.
Infiltrated inflammation cells, including myeloid and lymphoid immune cells, are important players in many normal organ and solid tumor microenvironments. The study depicted the distribution and functional characteristics of gastric tumor and adjacent normal tissue infiltrating myeloid cells. A few anti-inflammatory myeloid cell subtypes, including M07-APOE, M11-SPP1, M04-C3 are enriched in gastric tumor, suggesting an immune-suppressive microenvironment. In addition, such anti-inflammatory signature is significantly associated with reduced survival, further implicating the unfavorable influence of this immune-suppressive microenvironment. The lymphoid cells in gastric cancer microenvironment display similar transcriptional profiles compared to other cancer indications. The immune-suppressive Th17 and Treg populations both increase in the tumor tissue, reflecting the activation and expansion of T cells in the tumor.

Figure 3 Pro- and anti-inflammatory macrophages are negatively correlated and highly diverse between gastric cancer patients.
To investigate cellular interactions in the gastric cancer tumor microenvironment, the researchers constructed a cell communication network, and identified crucial cell populations and molecules from the network. The previously identified tumor-enriched stromal and myeloid cell types also exhibit important roles in the network. For example, the activated fibroblast F13-CTHRC1 acts as a central hub in the tumor microenvironment but not in the normal tissue. F13-CTHRC1 also interacts with a wide collection of other tumor-enriched cells and fosters an immune-suppressive stress through many ligands and receptors that are later validated to be upregulated in tumor compared to normal tissue. These findings align with each other in multiple dimensions and reveal an important tissue remodeling process in gastric tumorigenesis. In addition to communication between stromal and immune cells, some cells also directly interact with the malignant cells. A case is the malignant cell EP11-CLDN7, which inhibits the cytotoxic activity of CD8+ T cells through known immune checkpoint signals LGALS9-HAVCR2 and HVEM-CD160. EP11-CLDN7 also interacts with F13-CTHRC1 through the FZD5-WNT5A signal suggesting a potential role of F13-CTHRC1 to promote tumor cell migration.
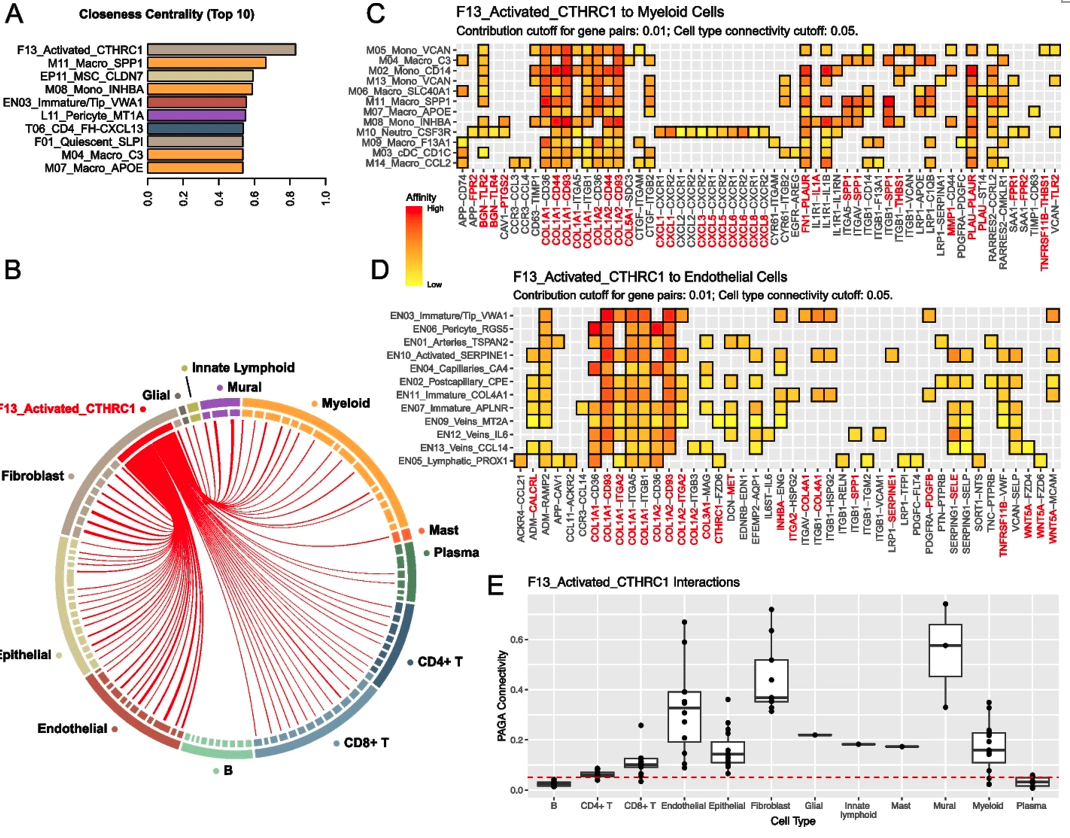
Figure 4 Cell communication inferred from single-cell transcriptome profiles in gastric cancer show a central role of F13-Activated-CTHRC1 fibroblasts.
Finally, the researchers used the carefully annotated cell expression signatures to analyze the TCGA and another independent anti-PD-1 treatment gastric cancer cohorts. The previously identified pro-tumor cell types are found to predict patient outcome in the TCGA cohort. Additionally, the signature of dendritic cell M16-cDC-CLEC9A is associated with better prognosis, suggesting their important role for antitumor immunity. In the anti-PD-1 cohort analysis, the signature T27-cycling CD8+ T cells followed by the presence of pan CD8 T cells are found to be a response predictor, whereas presence of F14-ADAM28 activated fibroblasts followed by presence of pan-fibroblasts are found to be a non-response predictor.
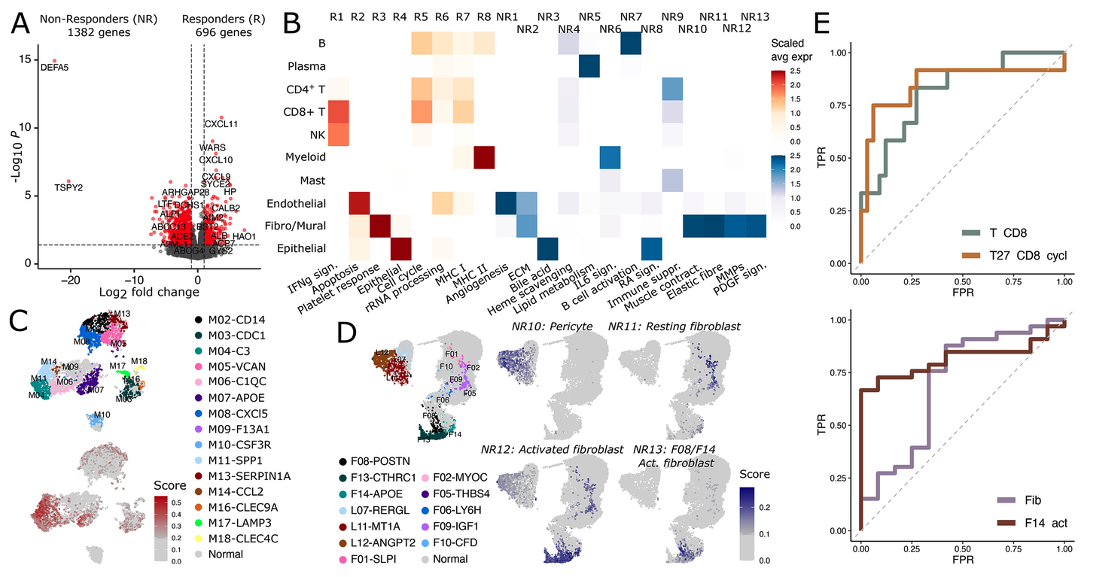
Figure 5 Cellular origin of response and non-response genes to cancer immunotherapy.
In summary, this study generated a multi-dimensional landscape for gastric tumor and adjacent normal microenvironments and identified multiple cell sub types and transcriptional programs that characterize the gastric cancer microenvironment. The important cell subtypes and molecules found in the study is helpful for developing new treatment strategies and biomarkers for patient stratification and increase the efficiency and efficacy for gastric cancer diagnosis and immunotherapy. The analysis methods employed by this study also provides a useful reference for other cancer and disease microenvironment research.
Dr. Boxi Kang of Peking University, Dr. Jordi Camps of Bayer AG, Dr. Biao Fan of Peking University Cancer Hospital, and Dr. Hongpeng Jiang of Peking University People’s Hospital are the co-first authors of this paper. Dr. Helge Roider of Bayer AG, Prof. Zemin Zhang of Peking University, Asso. Prof. Zhaode Bu of Peking University Cancer Hospital, and Prof. Zhanlong Shen of Peking University People’s Hospital are joint corresponding authors of this paper. This study is supported by National Natural Science Foundation of China, Beijing Municipal Science and Technology Commission, Science Foundation of Peking University Cancer Hospital, Beijing Advanced Innovation Center for Genomics, and Bayer AG. This study was approved by CHGRAO in July, 2019.
Link:
https://genomebiology.biomedcentral.com/articles/10.1186/s13059-022-02828-2