
Genome Biology | Systematic evaluation of colorectal cancer organoid system by single-cell RNA-Seq analysis
On April 28th, 2022, Tang Lab from Biomedical Pioneering Innovation Center (BIOPIC), Beijing Advanced Innovation Center for Genomics (ICG) collaborating with Wei Fu’s team from Peking University Third Hospital, published a paper entitled Systematic evaluation of colorectal cancer organoid system by single-cell RNA-Seq analysis on Genome Biology , which systematically evaluated the colorectal cancer organoid culture system.
Patient-derived tumor organoid systems have gradually become a promising cell culture system for cancer research. Although different types of cancer organoids have been wildly used in studying the molecular mechanism of tumorigenesis, the similarities and differences between cultured organoids and parental tumor tissues have not been systematically described at single cell level. And the transcriptome, DNA methylation and genome features of organoids cultured in different types of media remain unexplored. Here, the researchers systematically evaluated the molecular characteristics of colorectal cancer organoids and compared two common organoid culture media using high-precision single-cell RNA sequencing, whole-genome sequencing, whole-exome sequencing and targeted Sanger sequencing (Figure 1).
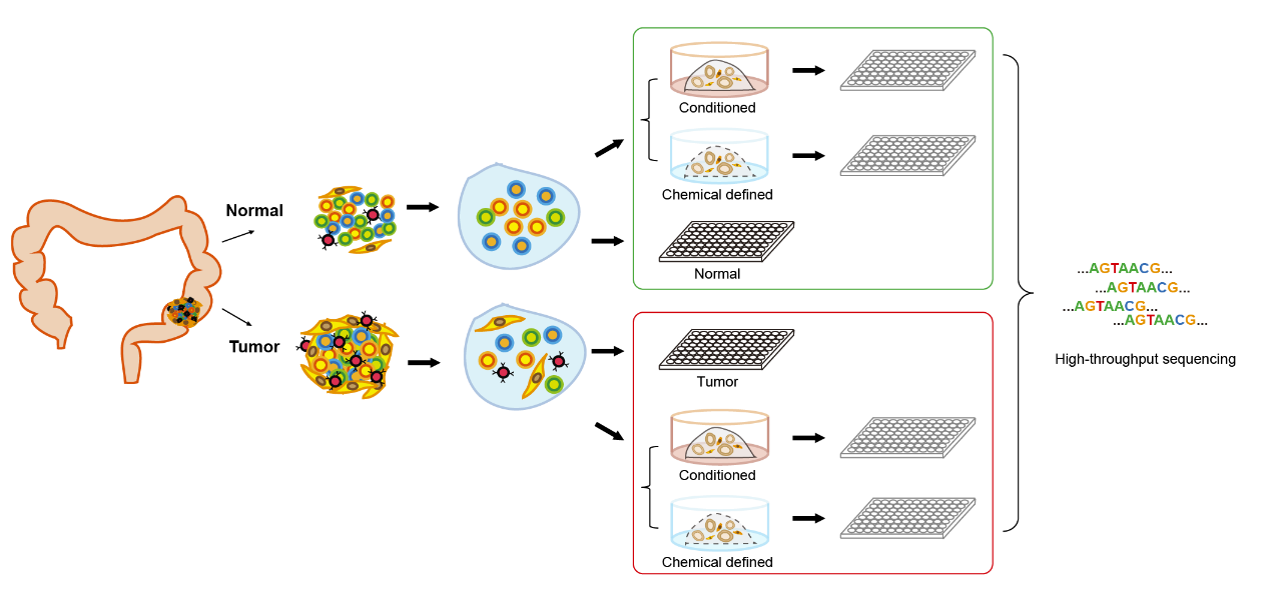
Figure 1 Experiment design
Tumor organoids accurately recapitulated genome, DNA methylation and transcriptome features of the corresponding tumor epithelial cells.
By exploring the single-cell transcriptome data, researchers found that tumor organoids faithfully recapitulated the gene expression and the gene regulatory network features of tumors in vivo. Moreover, organoids derived in both conditioned medium and chemical-defined medium well maintained the genomic features of tumor epithelial cells in vivo, such as copy number variation (CNV), gene mutation and global DNA methylation patterns (Figure 2). Colorectal cancer tumor organoids well recapitulated the key biological characteristics of cancer cells of corresponding patients, indicating that the organoid system is a valuable platform for studying colorectal cancer risk and drug development.
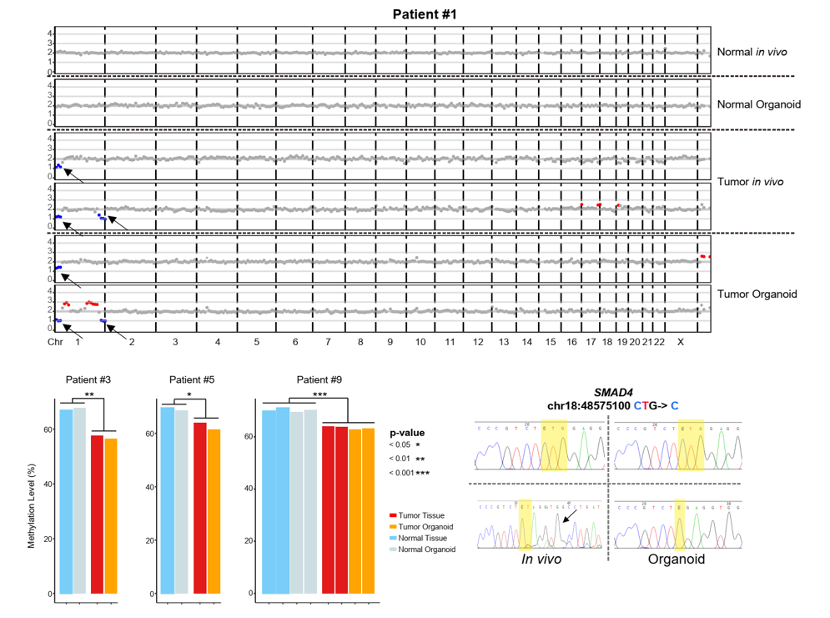
Figure 2 Comparison of the CNV; DNA methylation and the gene mutation features between cultured organoids and parental tissues
Normal tissue-derived organoids maintained the genomic and DNA methylation features of parental tissues, but exhibited some tumor-like features at the whole transcriptome level.
The researchers found that normal tissue-derived organoids exhibited tumor specific features at both transcriptome and protein level. For example, CEACAM6 was highly expressed in tumor tissues in vivo and corresponding organoids. However, the expression of CEACAM6 was also observed in normal tissue organoids. To further validate the results, researchers performed immunofluorescence staining. Consistent with single cell transcriptome data, both normal tissue-derived organoids and tumor organoids expressed CEACAM6 at protein level (Figure 3). To further exclude the potential contamination of tumor cells that might mingle in normal tissues, the researchers further investigated the genomic features of normal tissue-derived organoids and found that the normal tissue-derived organoids maintain the diploid and similar DNA methylation features of in vivo parental normal tissues. The results suggested that the existing normal tissue-derived organoids is not suitable as a control for the screening of anti-tumor drugs that selectively inhibit the tumor organoids and further improvement is required.
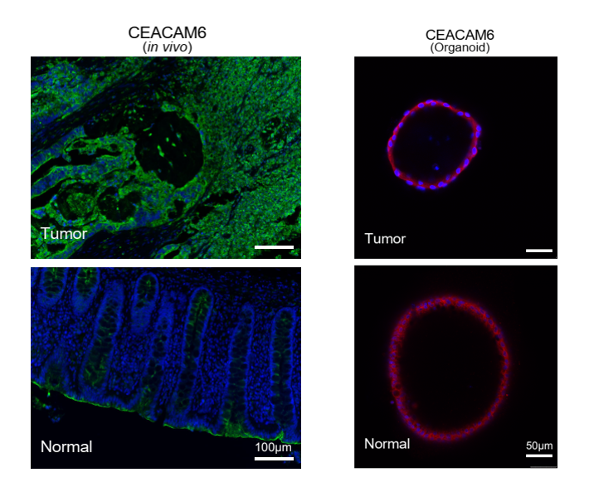
Figure 3 Immunofluorescence staining of CEACAM6
Conditioned medium outperformed chemical-defined medium during long-term culture of tumor organoids.
By evaluating cell proliferation rate in different media by the expression of MKI67, researchers found that normal tissue-derived organoids cultured in a chemical-defined medium had higher cell proliferation rates than tumor organoids. In contrast, the cell proliferation rates of normal tissue- and tumor-derived organoids were comparable in conditioned medium. This indicates that chemical-defined medium is more favorable for the growth of normal intestinal epithelial cells, whereas conditioned medium has no apparent preference for the growth of normal intestinal epithelial cells and tumor epithelial cells (Figure 4).

Figure 4 The ratio of cells that expressed MKI67.
Furthermore, organoids cultured in a conditioned medium maintained the differential expression patterns of enterocyte marker (CA2) and intestinal pluripotency marker (OLFM4) as the corresponding epithelial cells in vivo. However, in chemical-defined medium, tumor tissue-derived and normal tissue-derived organoid cells were mixed together, and there were no significant differences in the expression of CA2 and OLFM4, suggesting that chemical-defined medium favors stem cell-like features of normal epithelial cells whereas disfavors differentiation/mature features of them. These results indicated that although both conditioned medium and chemical-defined organoids can well maintain the genomic characteristics of parental tissues, the conditioned medium outperformed the chemical-defined medium and are the preferred culture system for organoid-based studies of molecular mechanisms of tumorigenesis and drug screening.
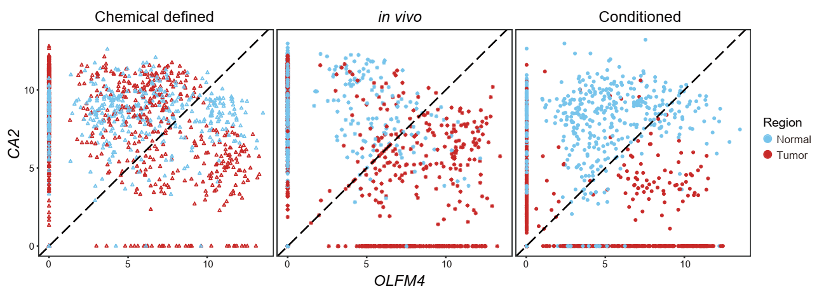
Figure 5 The expression levels of intestinal pluripotency marker OLFM4 and enterocyte marker CA2.
In summary, the research performed high-precision single-cell RNA sequencing of organoids cultured in two different types of media and the corresponding tissues. Combining whole-genome sequencing, whole-exome sequencing and targeted Sanger sequencing, the researchers have systematically assessed the reliability and limitations of the organoid culture system.
Dr. Rui Wang, Ph.D. candidate Yunuo Mao and Wei Wang from Peking University as well as Dr. Xin Zhou, Dr. Wendong Wang from Peking University Third Hospital are the co-first authors of the paper. Professor Fuchou Tang from Beijing Advanced Innovation Center for Genomics and Peking University and Professor Fu Wei from Department of General Surgery, Peking University Third Hospital are corresponding authors. This research project was supported by the National Natural Science Foundation of China and the Beijing Advanced Innovation Center for Genomics.
Link: https://genomebiology.biomedcentral.com/articles/10.1186/s13059-022-02673-3