
Nucleic Acids Research |Regulatory elements can be essential for maintaining broad chromatin organization and cell viability
On April 7, 2022, Prof. Wensheng Wei's group from Biomedical Pioneering Innovation Center (BIOPIC), Beijing Advanced Innovation Center for Genomics (ICG), School of Life Sciences in Peking University (PKU), collaborating with Prof. Wei Wang’s group from University of California, San Diego (UCSD), published a research article titled "Regulatory elements can be essential for maintaining broad chromatin organization and cell viability" in Nucleic Acids Research. Using the previously developed algorithm EpiTensor (Zhu et al. Nature Communications 2016) from Wei Wang’s group, the researchers identified genomic loci with active enhancer and promoter interactions named hotspots. Through high-throughput CRISPR/Cas9 library screening of hotspots by targeted deletion, dozens of noncoding loci were identified to be essential for cell growth and survival. This is the first report about the observations that deletion of such enhancers could alter a broad chromatin organization, and impact the expression levels of multiple individually non-essential genes concurrently, which exhibited synergistic effects to affect cell homeostasis.
In the noncoding regions of human genome, promoters and enhancers are important regulatory elements that control gene expression. In many cases, activated enhancers appear to engage in direct physical contact with their nearby promoters. However, there are also enhancers whose interacting promoters are distally located in the linear genome, and they are brought to spatial proximity by such as chromatin looping, protein oligomerization or Pol II tracking along chromatin, which highlights the important impact of 3D chromatin structure on the activities of these regulatory elements. Recently, an increasing number of studies have indicated that enhancer-promoter interactions play pivotal roles in forming specific 3D structures. Imaging analyses showed that transcription factors and polymerases are not evenly distributed in the nucleus but rather concentrated in certain regions associated with high transcriptional activities and a more compact chromatin structure. Transcription could also affect the 3D topology, such as transcription elongation was reported to be critical for chromatin organization. Whether these regulatory elements positioning in such spatial clusters with active transcription could contribute to maintaining broad chromatin structures therefore has become an emerging question.
To identify regulatory elements (promoters or enhancers) that are likely to be important for chromatin organization, researchers chose to start with those involved in many interactions with other genomic loci. As for the rare Hi-C data with sufficient resolution to define the interactions between promoters and enhancers, the researchers resorted to computational prediction by EpiTensor that can detect epigenetic covariation patterns between promoter-enhancer, promoter-promoter and enhancer-enhancer pairs at 200-bp resolution, and identified active promoters/enhancers that likely form many 3D spatial contacts with other active promoters or enhancers, referred to as hotspots hereinafter. They found that the regulatory element interaction networks (REINs) are small-world networks, which are characterized by robustness in that they are resistant to random attacks (random removal of nodes) but vulnerable to targeted attacks (removal of specific nodes) on high-degree nodes that have significantly more contacts than the other nodes (Figure 1A-B). To further investigate the functional roles of hotspots in maintaining chromatin organization, 751 hotspots identified as enhancers were randomly selected for targeted deletion through CRISPR/Cas9 library screening, 43 noncoding loci were identified as essential for cell growth and survival (Figure 1C). Among them, hotspot_10_25 (chr10: 74,123,469–74,1248,68) was validated to be specifically essential in K562 after deletion, while showed no significant growth defects in other five tested cancer cell lines (Figure 1D).

Figure1 Characterization of hotspot and the high-throughput functional screening
To investigated whether deleting hotspots affects chromatin organization, researchers selected hotspot_10_25 for further analysis, which does not interact with any essential protein-coding genes. Through the Hi-C analysis, they found that significant changes were observed on the 6–8 Mb regions surrounding the deleted hotspot for both effective diameter (chr10: 69–75 Mb for bin 71 and 72) and modularity score (chr10: 68–76 Mb for bin 70, 71 and 73). Some other genomic regions interacting with the hotspot neighboring regions were also affected, such as chr10: 11–17 Mb (bin 13 and 14), showing a significant change in modularity (Figure 2A-C). Consistently, we also found TAD splits in the region of chr10: 12–14 Mb with observed Hi-C contact changes. These chromatin changes led to alteration of promoter-enhancer interactions, such as the enhanced and weakened contacts between the CELF2, RSU1, FAM149B1 and CCAR1 promoters and their interacting enhancers upon hotspot deletion (Figure 2D). Notably, these affected promoters and enhancers are not only located close to but also can be as far as 62 Mb away from the deleted hotspot. These observations showed that hotspot deletion resulted in broad alterations in chromatin structure beyond its linear neighbor genome. Researchers further performed single-cell RNA-seq to analyse the changes in gene expression upon hotspot_10_25 deletion, and found apoptosis pathways are activated. Moreover, hotspot_10_25 deletion could affect the expression levels of multiple interacting genes located within the same TAD of the hotspot. Although none of these dysregulated genes was essential for cell fitness individually, altered expression of gene pairs showed significant synergistic effects leading to cell death (Figure 2E). This is the first report about the observations that enhancers could maintain a broad chromatin organization, which goes far beyond the direct interaction between promoters and enhancers.
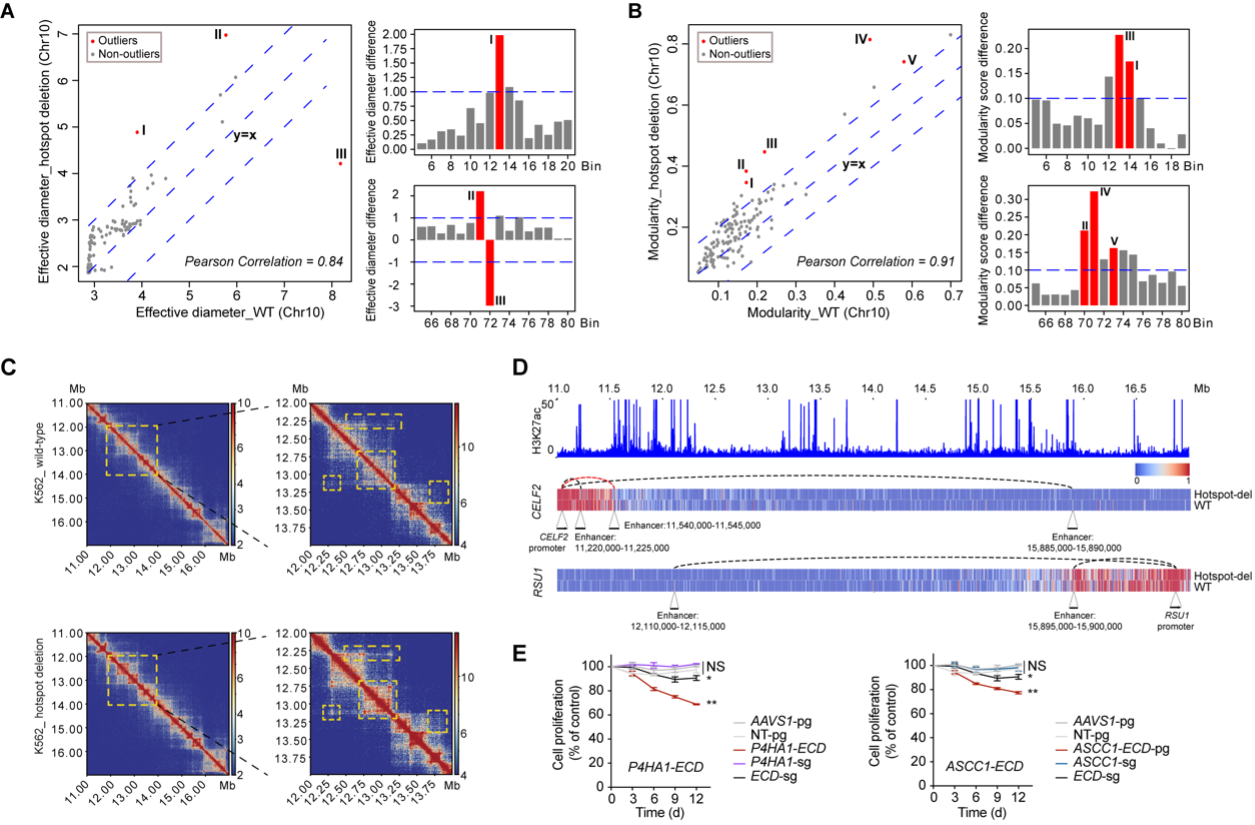
Figure 2 Deletion of an essential hotspot impacts broad chromatin structure and leads to synergistic change in gene expression
Notably, on November 2021, Science Advances published a research article titled "Noncoding loci without epigenomic signals can be essential for maintaining global chromatin organization and cell viability" from Prof. Wei Wang’s group (UCSD) and Prof. Wensheng Wei’s group (Ding et al., Science Advances 2021), reporting another category of noncoding loci named hub that are involved in chromatin structure. Through the systematic identification, high-throughput library screening and in-depth mechanistic analysis of the two categories of loci, the research will provide insights into exploring the biological functions and molecular mechanisms of the noncoding genome in the systems-level.
Dr. Ying Liu (PKU), Dr. Bo Ding (UCSD), Ph.D. candidate Lina Zheng (UCSD) and Dr. Ping Xu (PKU) are the co-first authors of the paper. Prof. Wensheng Wei and Prof. Wei Wang are the co-corresponding authors of the paper. This project was supported by funds from CIRM and the NIH (to Wei Wang); the National Science Foundation of China, Beijing Municipal Science & Technology Commission, the Beijing Advanced Innovation Center for Genomics at Peking University and the Peking-Tsinghua Center for Life Sciences (to Wensheng Wei); China Postdoctoral Science Foundation (to Ying Liu).
Article link:https://doi.org/10.1093/nar/gkac197