
Cancer Cell | Immune phenotypic linkage between colorectal cancer and liver metastasis
On March 17th, 2022, the Zhang Lab from Biomedical Pioneering Innovation Center (BIOPIC), Beijing Advanced Innovation Center for Genomics (ICG), School of Life Sciences in Peking University, collaborating with Dr.Peng from Beijing Shijitan Hospital, published a paper titled “Immune phenotypic linkage between colorectal cancer and liver metastasis” in Cancer Cell, which disentangled the factors shaping tumor microenvironment (TME) using the metastasis system.

The characteristics of TME are intrinsically associated with the efficacy of immunotherapeutic, and can significantly affect cancer progression and metastasis. Thus, it is essential to systematically dissect how cancer cells and host tissues affect the phenotype of different immune cells in the TME . However, their co-existence makes it difficult to disentangle their impacts on distinct TME components, as comparative analysis of TME between different cancer types cannot exclude the effects of different tumor cells from different patients. Unlike the cross-patients analysis, the process of tumor cells seeding the metastatic organ provides researchers with a unique system to unravel effects of niche and malignancy on TME, as metastatic tumors (seed) maintain genetic signatures from the primary tumor while migrating to a different organ (soil). Collecting different tumor loci from patients with metastatic tumor can help researchers resolve the TME formed by tumor cells with the same origin but in a different organ.
Researchers collected 101 matched samples from 17 treatment-naïve patients with colorectal cancer liver metastasis (CRLM), including primary colorectal cancer, adjacent colon, mesenteric lymph node, liver metastasis, adjacent liver and peripheral blood and analyzed the high-quality single-cell RNA-sequencing (scRNA-seq) transcriptome profile, generated by multi-platform. Further, 100 matched samples from 18 primary colorectal cancer (CRC) and 16 primary hepatocellular carcinoma (HCC) patients were integrated, and the PhenoAligner method was designed to evaluate the impacts of niche and malignancy on immune cell phenotypes (Figure 1).

Figure 1: Schematic diagram of the PhenoAligner method
Accordingly, the researchers identified immune populations whose phenotypes were significantly influenced by malignancy as M phenotype, immune populations whose phenotypes were significantly influenced by niche as N-type and those phenotypes influenced by multiple factors as C-type. The researchers systematically revealed the complex interactions between TME, cancer cells and organ microenvironment through building the linkage of the cellular components in the metastatic TME to their contributing tissue of origin (Figure 2).
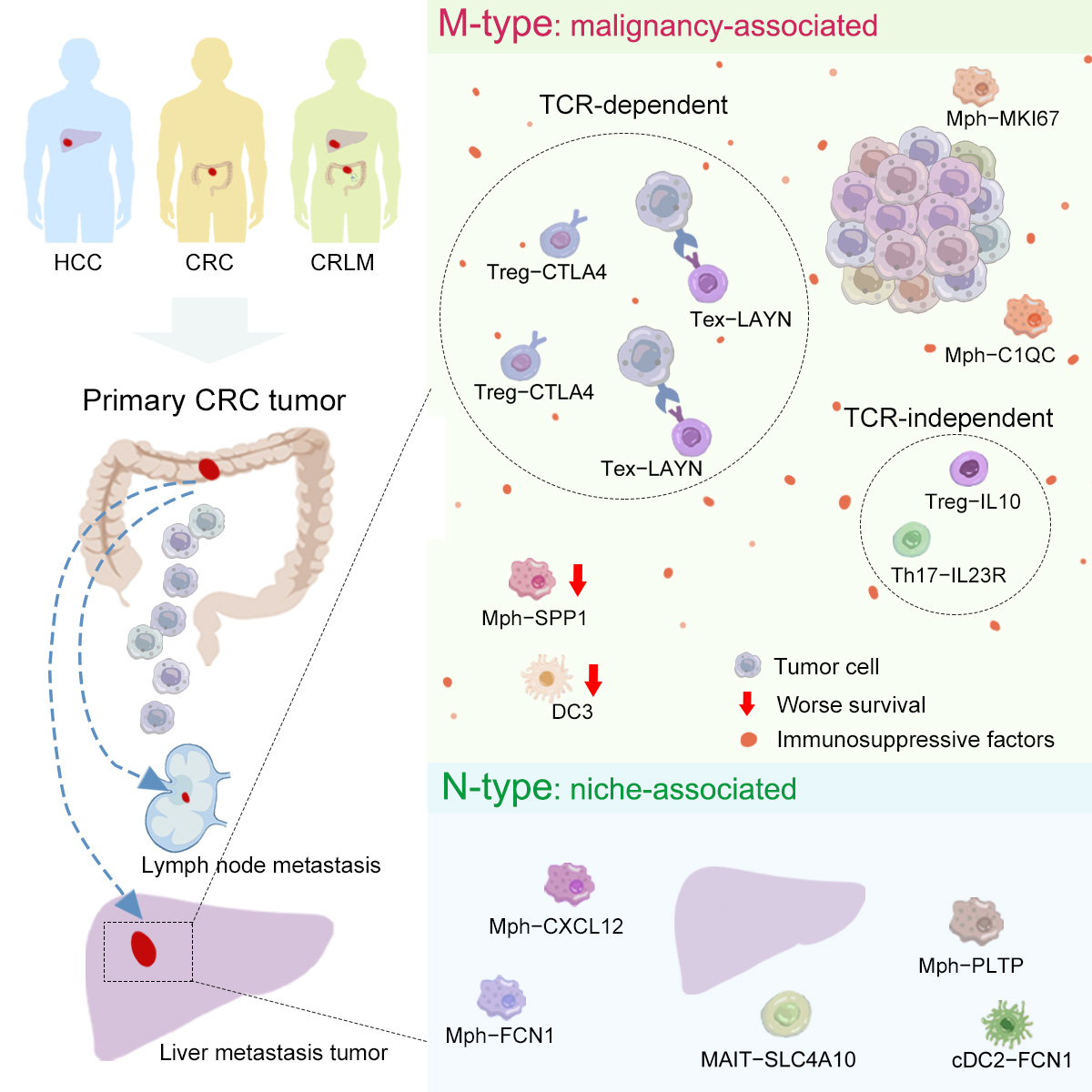
Figure 2: Scheme of the overall study design and major findings
The researchers classified exhausted CD8+ T cells (Texs) and activated Tregs as M-type, whose phenotype is associated with malignancy, while NK and MAIT cells were defined as N-type, whose phenotype is associated with the microenvironment. Due to the low mobility of Texs, the presence of identical TCRs between Texs in primary and metastatic tumors suggested they originated from the same precursor and triggered by cancer-associated antigens (Figure 3). The researchers came up with another mechanism in the following analysis of CD4+ T cells, that Th17 and Treg-IL10 cells exhibited mutually exclusive TCR usage in the two tumor sites while they were characterized as M-type.
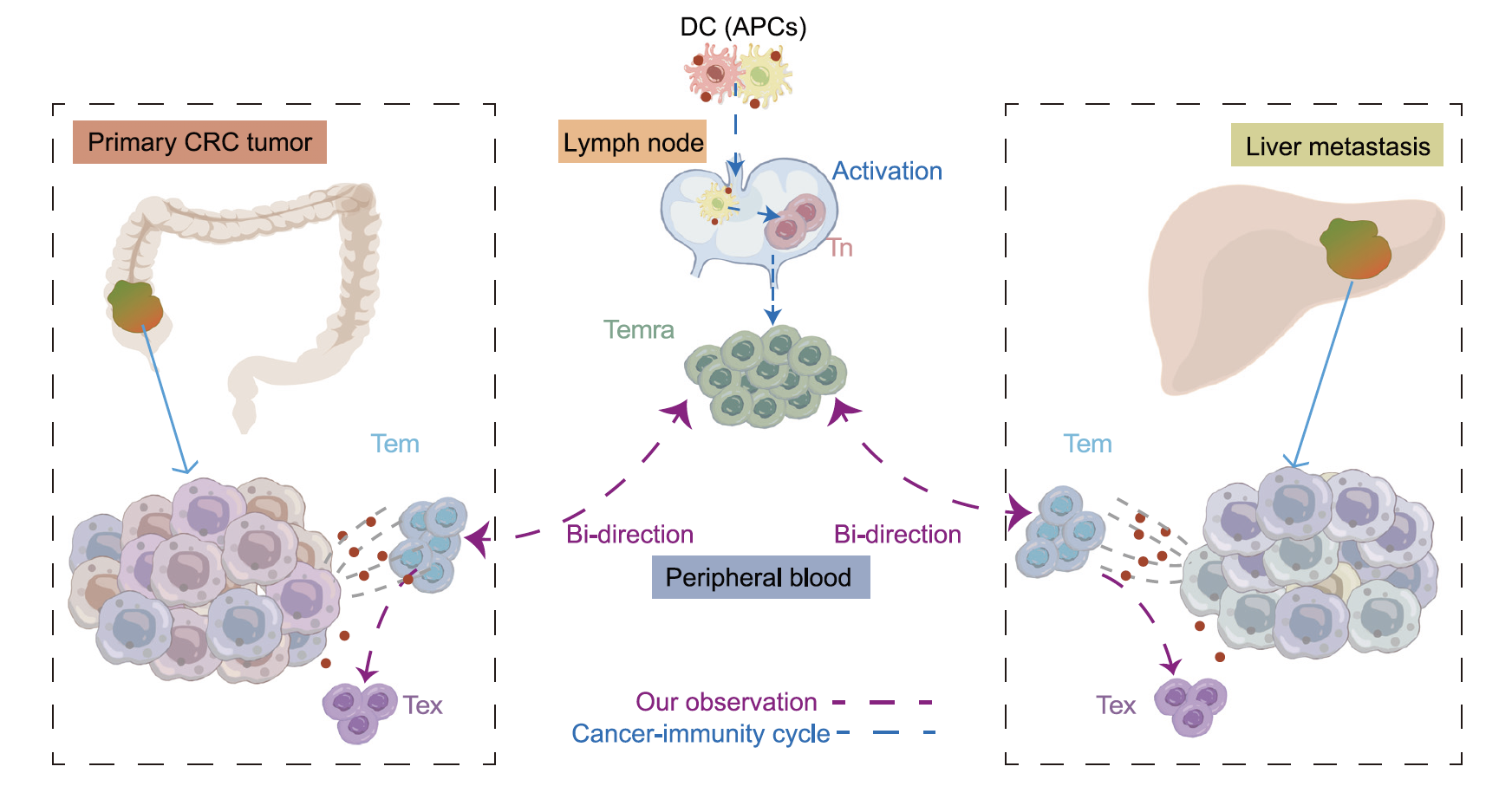
Figure 3: The process of CD8+ T cells infiltrating into different tumor sites
For myeloid cells, the researchers identified DC3s and SPP1+ macrophages as M-type, with the latter dominating in liver metastases, suggesting a pro-metastatic role. By comparing to the non-metastasis tumors, the researchers observed SPP1+ macrophages were absent in non-metastasis liver tumors, which further supported the possibility that malignancy are the primary driver of their phenotypes.
This metastasis model-based study linked the immune cell phenotypes between primary and metastatic tumors, helping to understand the specific immune features between distinct tumor types and to identify key components for the development of effective and low-toxicity immunotherapies.
Ph.D. candidate Yedan Liu and Qiming Zhang are the co-first authors of the paper. Prof. Zemin Zhang, associate Prof. Xianwen Ren and Prof. Jirun Peng are the co-corresponding authors of the paper. The project was funded by the National Natural Science Foundation of China, the Beijing Natural Science Foundation and the Beijing Advanced Innovation Centre for Genomics at Peking University.
Link: https://www.cell.com/cancer-cell/fulltext/S1535-6108(22)00065-4