
Nature Biotechnology|LEAPER 2.0: a circular RNA-based RNA editing tool with improved efficiency and fidelity
On Feb. 10th, 2022, the Wei Lab from Biomedical Pioneering Innovation Center (BIOPIC), Beijing Advanced Innovation Center for Genomics (ICG), and Peking-Tsinghua Center for Life Sciences (CLS) in Peking University, published a paper entitled Engineered Circular ADAR-Recruiting RNAs Increase the Efficiency and Fidelity of RNA Editing in Vitro and in Vivo in Nature Biotechnology , which describes an updated version of LEAPER termed LEAPER 2.0.
RNA editing is a trending technology that holds promise for treating disease without affecting genome integrity. In 2019, the Wensheng Wei group at Peking University reported in Nature Biotechnology a programmable RNA editing tool based on a human endogenous RNA editase. This technology dubbed “LEAPER” leveraged a specially designed RNA (ADAR-recruiting RNA, arRNA) to recruit the endogenous deaminase ADAR for making adenosine (A) → inosine (I) transition on target RNA【1】, circumventing the need for exogenous RNA editases, which may arouse aberrant immune responses and are commonly difficult for delivery to organisms.
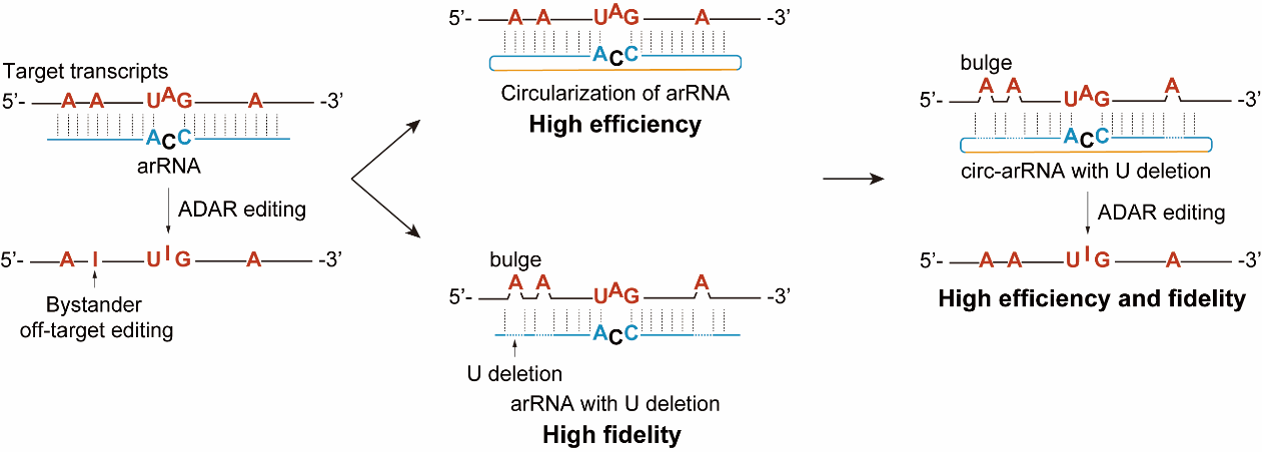
Because of its safety and simplicity, LEAPER demonstrated considerable potential in scientific research and medical treatment. However, it has limitations. First, LEAPER suffered from limited editing efficiency as it employs endogenous editing enzymes. Second, there are chances for ADAR to deaminate certain adenosines within the double-stranded region formed by targeting RNA and arRNA, resulting in bystander off-target editing. Therefore, there is still room for improvement in LEAPER′s efficiency and specificity.
Engineered circular ADAR-Recruiting RNAs increase the efficiency and fidelity of RNA editing.
In the paper, Yi et al . described an updated version of LEAPER termed “LEAPER 2.0”, in which they devised covalently closed circular arRNAs (circ-arRNA), instead of linear arRNA used in LEAPER, to recruit ADAR. This circ-arRNA, with significantly improved stability, achieved on average ~3-fold higher editing efficiency than their linear counterpart when expressed in cells or delivered as in vitro-transcribed circular RNA oligonucleotides. To lower off-target editing, they deleted uridines paired with potential off-target adenosines. This strategy eliminated nearly all bystander off-target editing. Engineered circ-arRNAs enhanced the editing efficiency and fidelity, as manifested by editing outcomes observed in endogenous CTNNB1 and mutant TP53 transcripts in cell culture. Delivery of circ-arRNAs using the adeno-associated virus in a mouse model of Hurler syndrome corrected the pathogenic point mutation and restored α-L-iduronidase catalytic activity, lowering glycosaminoglycan accumulation in the liver. As such, LEAPER 2.0 provides a new design of endogenous ADAR-recruiting RNA that enables more efficient and precise RNA editing with broad applicability for therapy and basic research.
Link: https://www.nature.com/articles/s41587-021-01180-3
1. Qu, L. et al. Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs. Nat Biotechnol 37, 1059-1069 (2019).