
Science | Pan-cancer single-cell landscape of tumor-infiltrating T cells
On Dec. 17th, 2021, the Zhang Lab from Biomedical Pioneering Innovation Center (BIOPIC), Beijing Advanced Innovation Center for Genomics (ICG), School of Life Sciences in Peking University, collaborating with Jiafu Ji’s and Zhaode Bu’s teams from Peking University Cancer Hospital, and teams from Peking University Third Hospital, published a research article entitled “Pan-Cancer Single-Cell Landscape of Tumor-Infiltrating T Cells” on Science. Combining gene expression profiles and T cell receptor sequences, the researchers investigated the heterogeneity and dynamics of tumor-infiltrating T cells and performed a systematic comparison of T cells among cancer types.

T cells play a central role in cancer immunotherapy, and cytotoxic T cells are the major subtype that could directly kill cancer cells. However, within the process of the tumorigenesis and evolution, these T cells usually differentiate into the dysfunctional state, which is widely known as T cell exhaustion. Immune checkpoint blockade (ICB) has shown tremendous clinical success, but its efficacy varies greatly across cancer types, which could be logically linked to the differences of tumor-infiltrating T cell states among cancer types.
Single-cell transcriptome sequencing (scRNA-seq) has been successfully applied to the analysis of the tumor microenvironment (TME) among a variety of cancers. For example, Zhang Lab and collaborators had previously performed T cells studies on three individual cancer types, including liver cancer, lung cancer, and colorectal cancer. However, there is still a lack of systematic comparison of tumor-infiltrating T cells across cancer types.
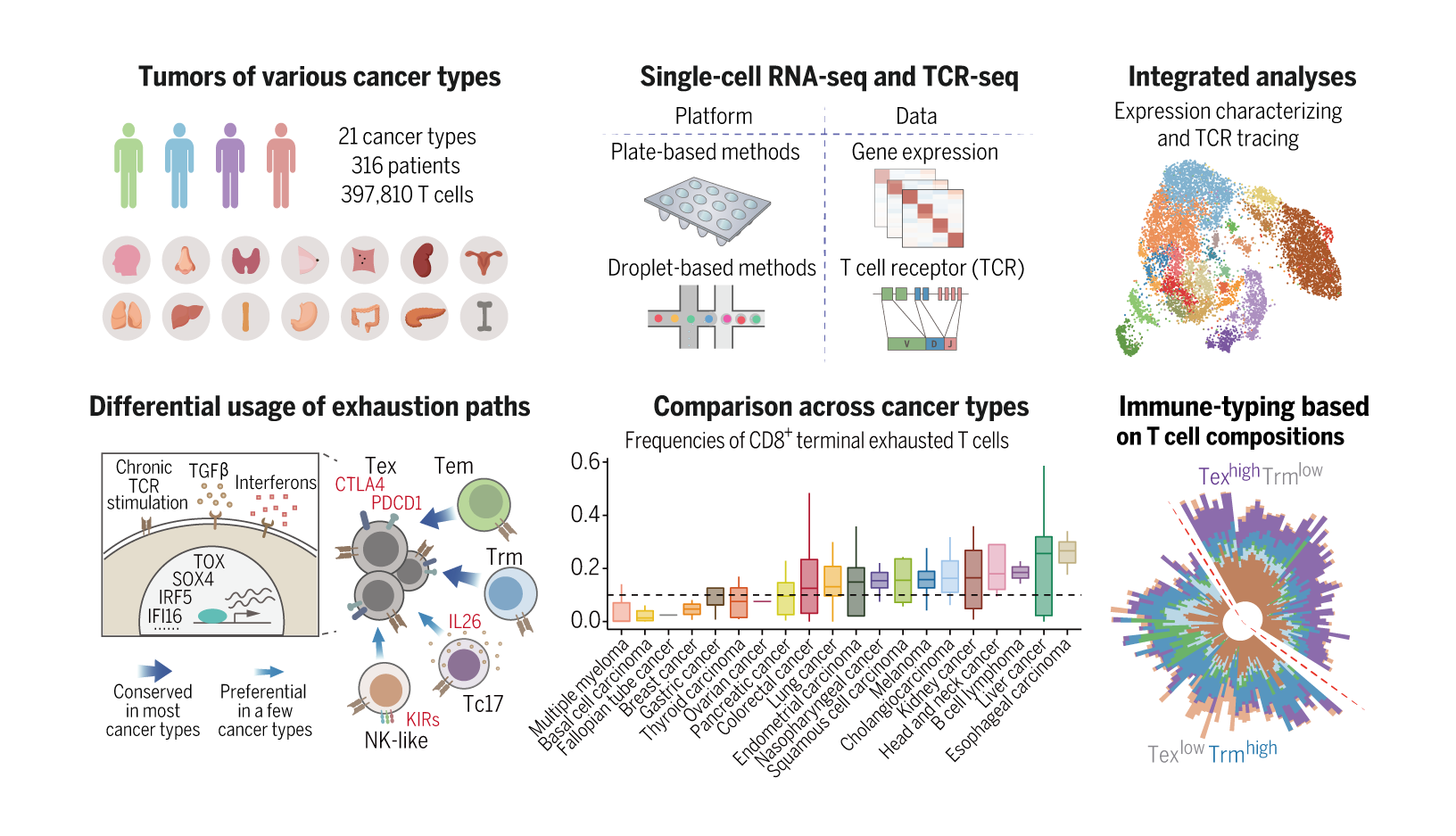
To better understand the landscape of tumor-infiltrating T cells and reveal the commonalities and differences of T cell states in different TMEs, the researchers have sequenced T cells for more cancer types, including myeloma, lymphoma, kidney cancer, ovarian cancer, endometrial cancer, esophageal cancer, thyroid cancer, breast cancer, gastric cancer, and pancreatic cancer, and also collected published data. The researchers utilized novel bioinformatics methods to correct confounding factors and batch effects, and integrated datasets from different platforms and sources. Thus, a high-resolution pan-cancer T cell atlas that contains 397,810 high-quality T cell data from 316 patients across 21 cancer types was constructed.
The researchers identified 17 CD8+ and 24 CD4+ T cell meta-clusters with distinct features in total and the compositions of T cells from different tissue-of-origin displayed prominent differences. Specifically, CD8+ T cells in the tumor were featured by the emergence of exhausted T cells, whereas among CD4+ T cell populations, the most abundant population was the TNFRSF9+ Treg cells. The results indicate that the TMEs have reshaped the states of T cells.
The researchers investigated the heterogeneity, dynamics, and transcriptional regulation of tumor-infiltrating T cells. In the CD8+ compartment, the researchers identified two major developmental paths to T cell exhaustion, respectively through effector memory T cells and tissue-resident memory T cells, and both were prevalent among cancer types. In addition, KIR+ NK-like T cells, Tc17 cells, or CD8+ Treg cells could also differentiate into exhausted T cells, but these transitions were only observed in a few cancer types. The researchers further analyzed the transcription factors (TFs) associated with T cell exhaustion. TFs such as TOX and PRDM1 were highly expressed in exhausted T cells in most cancer types, but certain TFs showed preference in a few cancer types, such as SOX4 and FOXP3. In the CD4+ compartment, the major potentially tumor-reactive T cells were IFNG+ TFH/TH1 and TNFRSF9+ Treg cells. The researchers inferred that IFNG+TFH/TH1 cells may be differentiated from classic IL21+TFH cells. TNFRSF9+ Treg cells were mainly derived from TNFRSF9- resting Treg cells, but also exhibited certain state transition potentials with non–Treg cells such as TH17 and TFH in a few cancer types.
The researchers also examined extrinsic factors associated with T cell compositions in the tumor. For example, the tumor mutation burden (TMB) showed a positive association with TFH/TH1 cells, and FAT1 mutations exhibited a positive correlation with TNFRSF9+ Treg cells. Cancer types exerted an extensive impact on the frequencies of T cell populations. Moreover, the researchers proposed to stratify patients based on the composition of tumor-infiltrating T cells. The patients could be divided into two groups — one with a higher proportion of terminally exhausted CD8+ T cells and the other one with a higher proportion of tissue-resident memory CD8+ T cells. The latter group has better survival than the former.
This study provides new insights into T cell immunity and will promote the development of new strategies targeting T cells to treat human cancer.
Systematic analysis of a human pan-cancer T cell atlas
Dr. Liangtao Zheng, Ph.D. candidate Shishang Qin, and Dr. Wen Si are the co-first authors of the paper. Drs. Zemin Zhang, Jiafu Ji, Zhaode Bu, and Dr. Xueda Hu (from Analytical Biosciences) are the co-corresponding authors of the paper. The project was funded by the National Natural Science Foundation of China, the Beijing Natural Science Foundation, and the Beijing Advanced Innovation Centre for Genomics at Peking University, and supported by the Computing Platform of the Center for Life Science, Peking University.