
PLoS Biol. ∣ Dr. Lu Wen, Dr. Lei Song and Dr. Jizheng Wang Reveal Multiomics Map of Human Hearts
The heart, which is composed of 4 chambers, is the first organ to be formed during mammalian development. Heart is physically essential from embryonic to adult stages, and heart diseases are one of the No. 1 killers of human health. Current heart researches have mainly separately analyzed human or mouse, but the species difference between human and mouse hearts, with simutaneous analyse on the chamber-specific characteristics, have not yet been comprehensively investigated.

On May 18, 2021, Associate Research Scientist Dr Lu Wen in Biomedical Pioneering Innovation Center (BIOPIC) of Peking University, in collaboration with Professor Lei Song and Dr Jizheng Wang of Fuwai Hospital, published an article entitled Integrated transcriptomics and epigenomics reveal chamber-specific and species-specific characteristics of human and mouse hearts online in PLOS Biology . This study collected 11 adult human hearts from healthy donors, 3 fetal human hearts, 3 groups of male C57BL/6 mice and performed NOMe-seq and ribosomal RNA-depleted RNA-seq for them, which revealed chamber-specific and species-specific characteristics of gene expression, DNA methylation, and chromatin accessibility of human and mouse hearts.
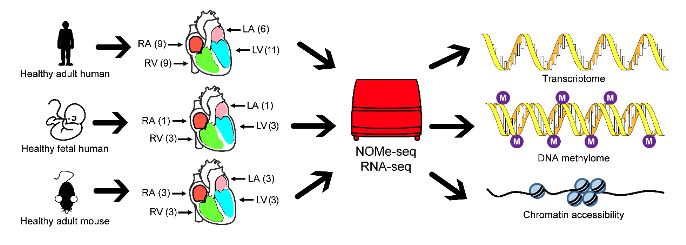
Figure 1 Experimental design of the study.
The study found that there was significant difference between the atria and the ventricles of the adult human heart in transcriptome and DNA methylation. They identified a series of differentially expressed genes and several genes that showed different methylation levels in gene-body regions between the atria and the ventricles (Figure 2A). Considering chromatin accessibility, there were no significant differences in the overall chromatin accessibility in the 4 chambers of the heart. When comparing fetal and adult human hearts, they found there were obvious chamber-specific and developmental stage-specific characteristics in transcriptome and DNA methylation (Figure 2B), and developmental stage-specific characteristics in chromatin accessibility. 176 novel lncRNAs were identified in human heart and they showed different expression pattern in 4 chambers of adult and fetal heart (Figure 2C). The regional and temporal specificity of cardiovascular disease-associated genes demonstrated that the physiology and pathology of the human heart are under precise spatiotemporal expression and regulation.
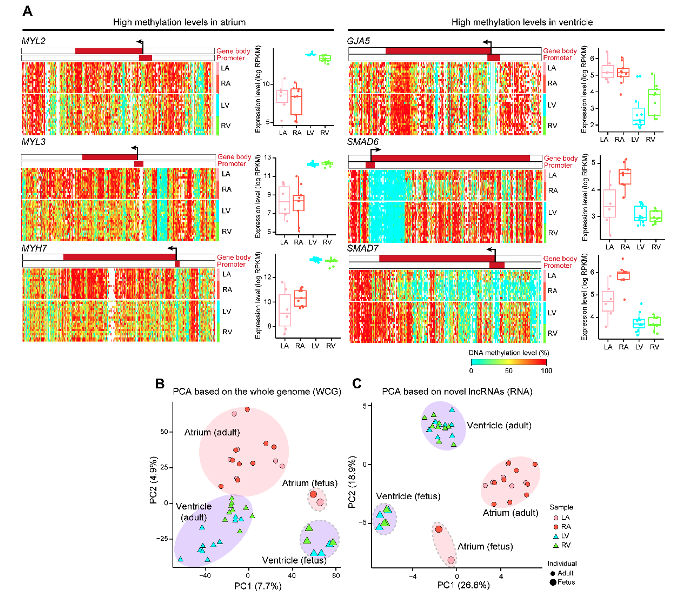
Figure 2 Heatmaps showing endogenous DNA methylation levels around the gene body ± 10 kb regions (left) and boxplots showing the gene expression levels (right) of genes in the adult human heart.(A);PCA plot showing the endogenous DNA methylation pattern of the whole genome (B) and the novel lncRNAs (C).
The mouse is the most widely used model for studying heart development and cardiac diseases. Although human and mouse hearts are similar in anatomy, they are quite different in size and electrophysiology. Additionally, studies have shown that human and mouse hearts respond differently to cardiovascular drugs. So it is necessary to compare human and mouse hearts in different levels. This study found that human and mouse hearts shared some important DEGs between the atria and the ventricles (Figure 3). Similar to human heart, mouse heart showed obvious chamber-specific pattern. Myl2 and Myl3 showed gene-body hypomethylation in the ventricles in both human and mouse heart. Our data showed that the NDRs of human and mouse hearts shared many TF motifs, indicating that the main regulatory networks are conserved between these 2 species.

Figure 3 (A) Scatter plots showing fold change of the overlapping DEGs (protein-coding genes) in cross-species comparisons. (B) GO terms of human- (left) and mouse- (right) specific DEGs.
This study also investigated correlations among DNA methylation, chromatin accessibility, and gene expression. They found a negative correlation between gene expression and DNA methylation with positively correlated gene expression and chromatin accessibility in human heart (Figure 4A), which was reported by previous study. Indeed, gene-body methylation displayed a bell-shaped relationship with gene expression, with high-expression genes showing prominently lower methylation levels than intermediate-expression genes (Figure 4B). The detailed mechanism underlying the bell-shaped relationship between expression and gene-body DNA methylation or chromatin accessibility needs further research in the future.
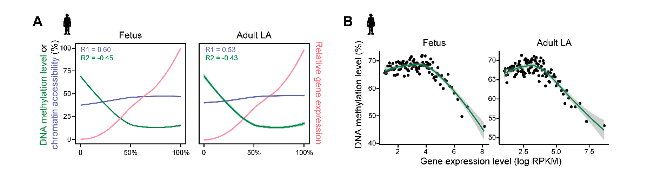
Figure 4 Correlations among multiomics of human heart in promoter regions (A) and gene-body regions (B).
In conclusion, this study provides integrated transcriptome, DNA methylation, and chromatin accessibility maps for understanding the spatiotemporal characteristics of both the human and mouse hearts. The resulting multiomics map of healthy human and mouse hearts can be used as a reference to identify novel biomarkers or drug targets when compared with malfunctioning hearts.
Ph.D. candidate Junpeng Gao from Biomedical Pioneering Innovation Center (BIOPIC) and Yuxuan Zheng from Academy for Advanced Interdisciplinary Studies of Peking University are the co-first authors of the paper. Research Scientist Dr Lu Wen of Beijing Advanced Innovation Center for Genomics, Biomedical Pioneering Innovation Center, School of Life Sciences of Peking University, and Professor Lei Song and Dr Jizheng Wang from Fuwai Hospital are the co-corresponding authors. This research project was supported by the National Natural Science Foundation of China and the Beijing Advanced Innovation Center for Genomics.
Link: https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3001229