
Nature Communications| Single-cell transcriptomics links malignant T cells to the tumor immune landscape in cutaneous T cell lymphoma
On Mar. 3rd, 2022, the Fan Bai Group from Biomedical Pioneering Innovation Center (BIOPIC) and Beijing Advanced Innovation Center for Genomics (ICG) in Peking University, collaborating with Yang Wang Group at Peking University First Hospital, published an article entitled “Single-cell transcriptomics links malignant T cells to the tumor immune landscape in cutaneous T cell lymphoma” in Nature Communications. This study characterized cutaneous T cell lymphoma (CTCL) and their immune microenvironment at the single-cell level, proposed a multi-step tumor evolution model and established a subtyping scheme based on the molecular features of malignant T cells and their pro-tumorigenic microenvironments.

Cutaneous T cell lymphomas (CTCL) is a heterogeneous group of extranodal non-Hodgkin’s lymphomas characterized by cutaneous infiltration of clonal malignant T cells [1]. CTCL generally exhibits an indolent course, but a portion of patients progresses rapidly and exhibit widespread disease beyond the skin despite aggressive treatment, highlighting the underlying heterogeneity of this disease [2-3]. Moreover, CTCL patients frequently show inter-lesional diversity in treatment responses, adding to the complexity of the clinical challenge of the disease and often creating obstacles to achieving effective therapy [4]. Elucidating how malignant T cells redefine and alter the tumor microenvironment (TME) and clarifying the landscapes and properties of the multicellular ecosystem in CTCL will create new opportunities for the development and application of immunotherapies.
With single-cell RNA sequencing (scRNA-seq) and paired single-cell T cell receptor (TCR) sequencing on 19 skin lesions from 15 CTCL patients, malignant T cells were defined by copy number variations (CNVs) and matched TCRα and TCRβ clonotypes at the single-cell level (Fig. 1). With this approach, the researchers determined the mono-clonal nature of CTCL and characterized the temporal and topological subclonal evolutionary process of malignant T cells, revealing a high degree of tumoral cellular heterogeneity. The genetic basis of this intratumor heterogeneity (ITH) was defined by paired whole-exome sequencing (WES). The researchers generated a molecular subtyping scheme based on the intrinsic identity of malignant T cells across CTCL variants and deciphered the complex crosstalk among immune cells within the TME for each subtype.
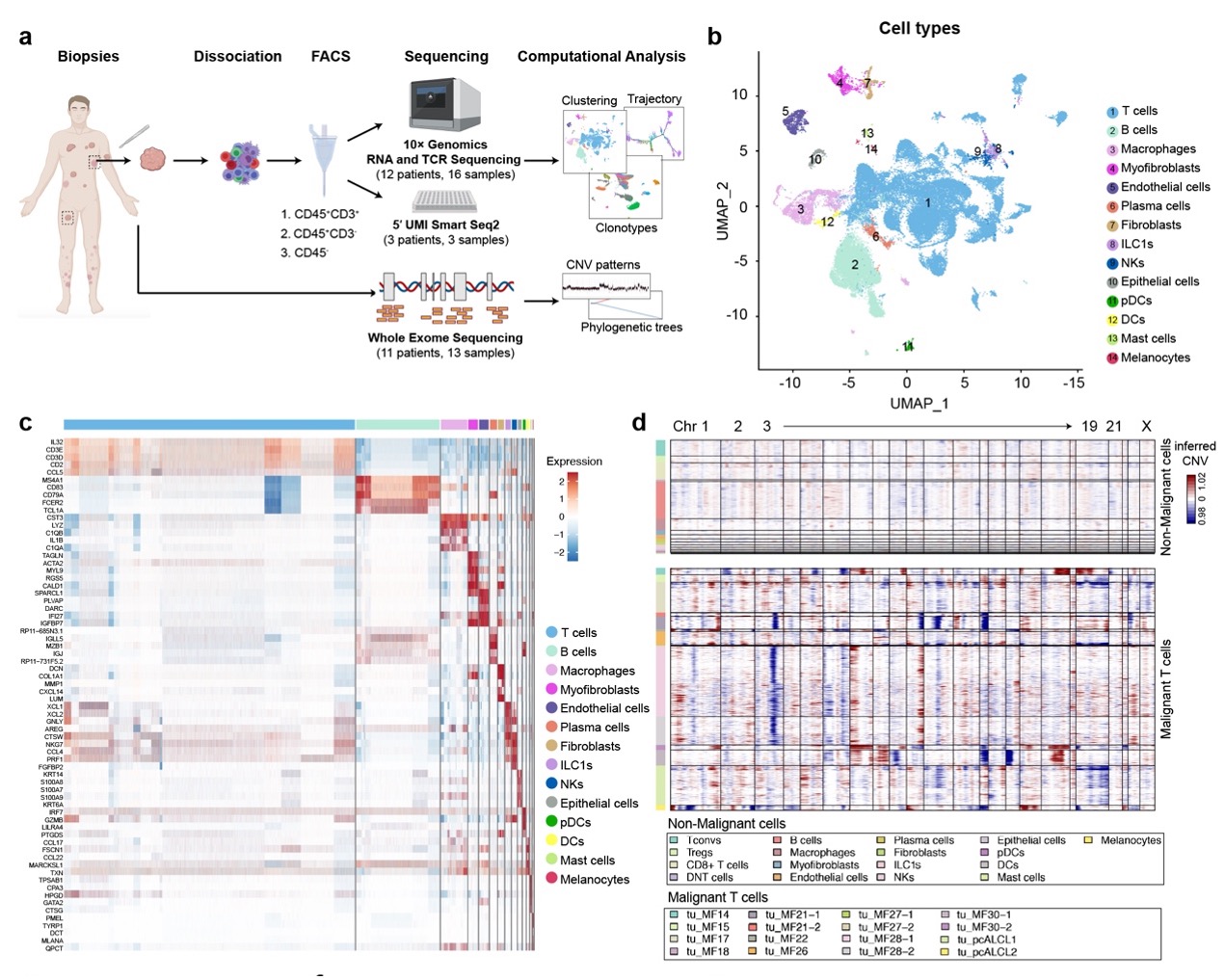
Fig. 1 Single-cell transcriptional profiling of 19 cutaneous T cell lymphoma samples
To explore the temporal and spatial spreading patterns of CTCL, the researchers analyzed paired tumors obtained from different anatomical sites from 3 CD4+ MF patients (patients MF21, MF28, and MF30). Interestingly, the transcriptome heterogeneity increased along with the time interval of lesion development and the anatomical distance between lesions in individual patients. Accordingly, the researchers proposed a multi-step tumor evolution model (Fig. 2a): These two tumors obtained from different anatomical sites arose from an early clone that was seeded to the skin of the entire body in the early stage of the disease, possibly before clinically detected lesions appeared, and subclonal tumor evolution subsequently occurred in parallel in the skin. The subclonal malignant T cells in a well-developed tumor may re-circulate and seed into adjacent skin areas and generate late-arising lesions. This dynamic evolution leads to the accumulation of ITH within each skin lesion, as well as inter-lesion diversity, and contributes to the heterogeneous clinical features and treatment response observed among different skin lesions in the same patient.
Meanwhile, the researchers established a molecular subtyping scheme in CTCL based on the transcriptome of malignant T cells (Fig. 2b-c). The researchers defined a TCyEM group, in which the malignant T cells showed an activated cytotoxic effector memory T cell phenotype, as well as a TCM group, in which malignant T cells were central memory T cell-like cells with high proliferation and exhaustion status. Compared to the TCyEM group, the molecular features of TCM patients were related to more advanced disease stages, aggressive behavior, and adverse prognosis. This subtyping may represent the distinct origins of malignant T cells and are related to the patient outcome as assessed in a larger cohort (n="49).
Malignant TCyEM cells appeared to produce high CCL5 and CCL4 to recruit and polarize CD163-expressing M2 TAMs, forming a tumor-supporting environment in TCyEM cases. Global down-regulation of MHC-I molecules and up-regulation of galectin 9 collaboratively contributed to tumor evasion from cytolytic CD8+ TIL-mediated antitumor immunity in the TCM group. And malignant T cells activated B cells in TCM patients via a CD40L/CD40 axis, and highly infiltrated B cells may promote tumor progression by activating regulatory T cells (Tregs). The researchers showed that the origins of malignant T cells determined the particular characteristics of their distinct TME, which could be exploited to develop tailored immunotherapy strategies for different subtypes of patients.
Collectively, this work establishes a conceptual foundation for understanding CTCL tumorigenesis and proposes a molecular subtyping strategy incorporating the phenotypes of malignant T cells and the multicellular immune landscape.
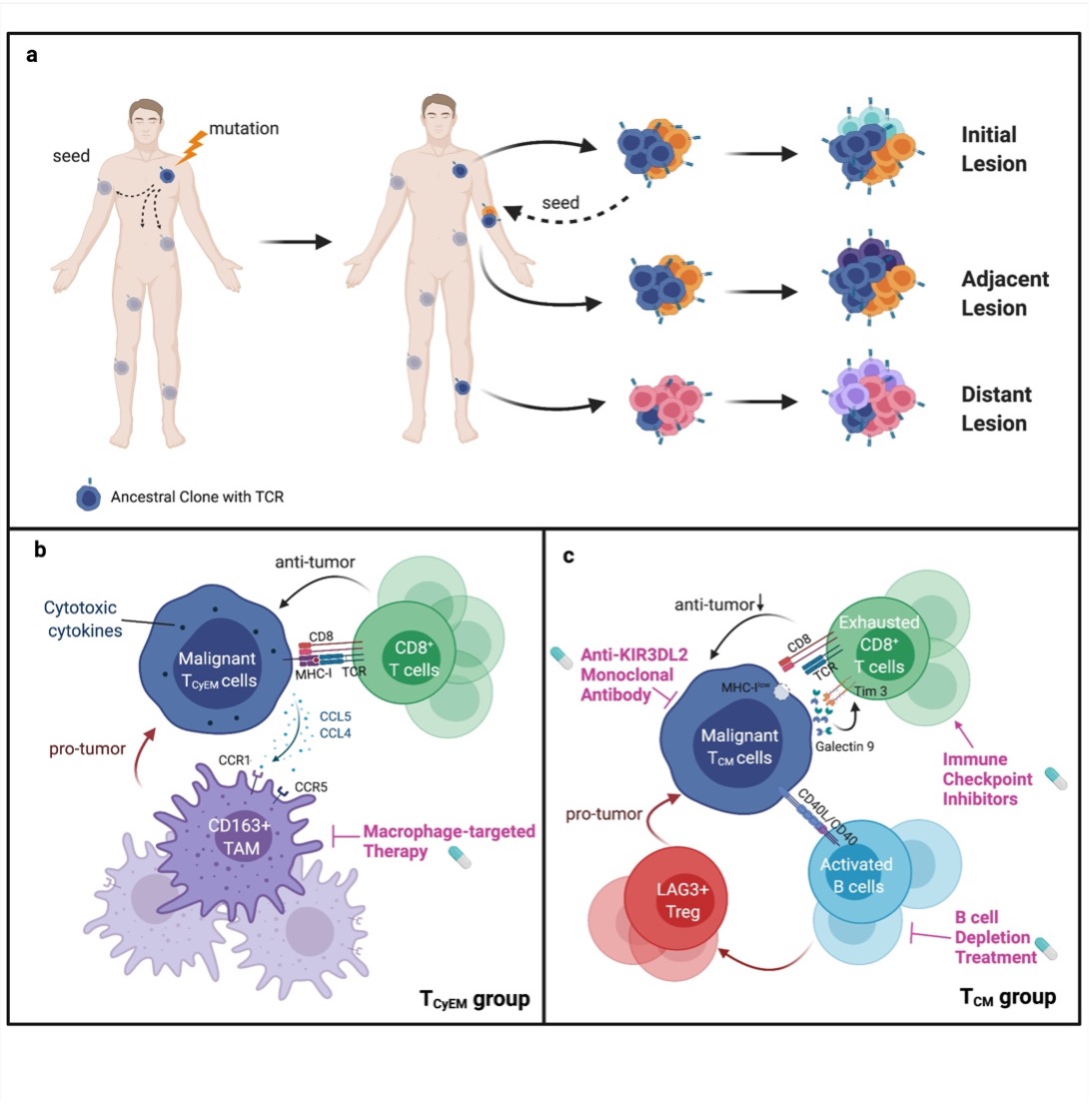
Fig. 2 A multi-step seeding model and molecular subtyping schematic representation of this study.
Dr. Xiangjun Liu, Dr. Shanzhao Jin, and Ph.D. candidate Simeng Hu are the co-first authors of the paper. Prof. Yang Wang from Peking University First Hospital and Prof. Fan Bai from Peking University are the co-corresponding authors of the paper. This research was supported by the National Nature Science Foundation of China, National Science and Technology Major Project, Guangdong Province Key Research and Development Program, National Youth Top-Notch Talent Support Program, and Peking University Clinical Medicine plus X Youth Project.
References:
[1] Criscione et al., Incidence of cutaneous T-cell lymphoma in the United States, 1973-2002, Arch Dermatol (2007).
[2] Arulogun et al., Long-term outcomes of patients with advanced-stage cutaneous T-cell lymphoma and large cell transformation. Blood (2008).
[3] Iyer et al., Branched evolution and genomic intratumor heterogeneity in the pathogenesis of cutaneous T-cell lymphoma. Blood Adv (2020).
[4] Willemze et al., WHO-EORTC classification for cutaneous lymphomas. Blood (2005).