
Science | Single-Cell Multi-Omics Technology Revealing the Relationship Between the 3D Genome Architecture and Gene Expression During Development
In higher eukaryotes, the three-dimensional (3D) chromatin structure within the cell nucleus plays a vital role in cellular function. For example, enhancers often regulate the transcriptional activity of distal target genes through 3D spatial interactions 1-3. Abnormalities at the 3D genome level have also been found to be closely associated with the occurrence of various diseases, including cancer 4-6. However, the overall relationship between the 3D genome and gene expression remains controversial. For instance, significant rearrangements in the 3D genome can occur through the targeted degradation of key regulatory proteins such as CTCF or cohesin, but this only results in a minor impact on gene expression 7-9 . In Drosophila embryos, there are significant differences in gene expression between different cell types, but the chromatin structure differences are not prominent 10,11. Therefore, to further understand the relationship between 3D chromatin structure and gene expression under the context of complex tissue organs and diverse cell types, more powerful tools are urgently needed.
On June 9, 2023, a research group led by Dong Xing from the Biomedical Pioneering Innovation Center (BIOPIC) of Peking University published a research paper in Science titled "Linking genome structures to functions by simultaneous single-cell Hi-C and RNA-seq". They reported a novel single-cell multi-omics technique called HiRES (Hi-C and RNA-seq employed simultaneously), which, for the first time, enabled simultaneous detection of the transcriptome and 3D genome at the single-cell level based on sequencing methods.
The HiRES method firstly performs in situ reverse transcription and chromatin conformation capture (3C) at the population level, and then obtains single cells through flow sorting. Each single cell is then amplified and sequenced (Fig. 1A). DNA or RNA reads are distinguished by the RNA recognition sequence introduced during the reverse transcription process, thus the method does not involve physical separation of DNA and RNA, maximizing the detection efficiency. In mouse brain samples, an average of 6,517 genes, 27,468 transcripts (UMIs), and 317,435 chromatin interactions could be detected per cell (Fig. 1B, C). With this data, researchers reconstructed the high-resolution structure of the single-cell 3D genome, and mapped the allelic resolved gene expression profiles on the 3D structure (Fig. 1D, E).
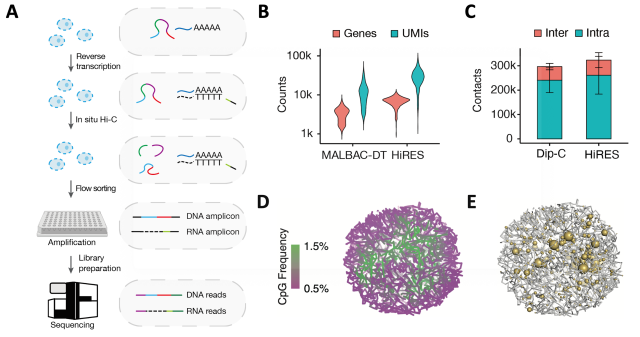
Figure 1 HiRES simultaneously profiles 3D genome structures and gene expression with high efficiency and accuracy. (A) Schematics of the HiRES workflow. (B) Number of genes and RNA unique molecular identifiers (UMIs) for MALBAC-DT and HiRES. (C) Number of chromatin contacts for Dip-C and HiRES . (D) Example of a single-cell 3D genome structure colored by A/B compartmentalization. Compartmentalization of euchromatin (green) and heterochromatin (magenta) was visualized by cytosine-phosphate-guanine (CpG) frequency as a proxy. (E) Example of 3D genome structure rendered together with gene expression. The size of the yellow balls is proportional to the transcription level.
Following this, using the HiRES method, the researchers mapped the transcriptome and 3D genome of a total of 7,469 single cells from mouse embryos from day 7 to 11.5 post-fertilization. The analysis of this dual-omics data allowed the research to explore the following aspects:
1.Cell-cycle dynamics in different cell types
During embryonic development, cells are in a process of rapid proliferation, and the influence of the cell cycle on the three-dimensional genome cannot be ignored. This study took advantage of the double-modality data of HiRES and developed a strategy to assign single cells to different cell-cycle phases. This strategy mainly relies on indicators such as DNA spatial interaction characteristics, cell cycle-related gene expression, and DNA replication, which can assign single cells to seven different cell-cycle states (Figure 2).
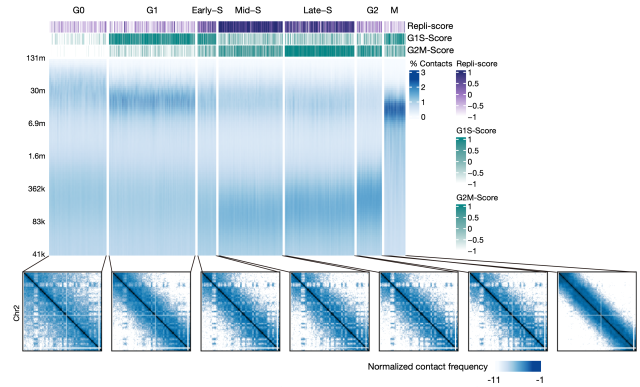
Figure 2 Cell-cycle phases annotation with dual-omics data. Each column of the heatmap represent a single cell.
In embryonic tissues, the cell clusters defined by chromatin structure were more consistent with cell-cycle states, suggesting chromatin conformational changes coordinated by cell cycle as a major contributor to the overall 3D genome structure. Between different cell types, this study developed a semi-quantitative method to compare the chromatin structures during the cell cycle process. It was found that the interphase cells from the late developmental stage have more fully established long-distance interactions after mitosis, while the differences between early and late-stage cells gradually decrease with the progress of DNA replication. These results indicated that chromatin organization of interphase cells was determined by two independent processes, the spontaneous unfolding of chromosomes after mitotic exit and the replication of the genome. For cell cycles with a short G1, the two processes overlapped in time and together determined the cell-cycle dynamics of the 3D genome.
2. The differences in chromatin interactions among different cell types are closely related to cellular functions.
Investigating the chromatin structural differences of different cell types is one of the major issues in 3D genomic research. Using bulk Hi-C to find differential chromatin interactions (DIs) is challenging because of the high technical noise caused by proximity ligation and the limited number of biological replicates. In addressing these issues, this study designed the SimpleDiff strategy. By conducting a non-parametric test on the spatial distance between two points in the reconstructed whole-genome three-dimensional structure of two groups of single cells, significant DIs can be efficiently detected. Based on this method, the study found that the specialization of chromatin structure has appeared during gastrulation, and DIs significantly enriched differentially expressed genes, indicating that changes in chromatin structure are closely related to cell function. On this basis, the study further connected differential interactions with their potentially related genes by utilizing the covariation of in chromatin interactions and gene expression. Differential interactions significantly correlated with gene expression were referred as GADI (gene-associated DI).
The study found that GADIs often link promoters to distal regulatory elements (such as enhancers, super-enhancers, Figure 3), many of which contain regulatory elements whose functions have been identified through in vitro CRISPR knockout experiments. Therefore, GADIs not only contain the results of transcriptional activity but also include many potential enhancer-promoter interactions, which are the basis for cell type-specific gene expression control.
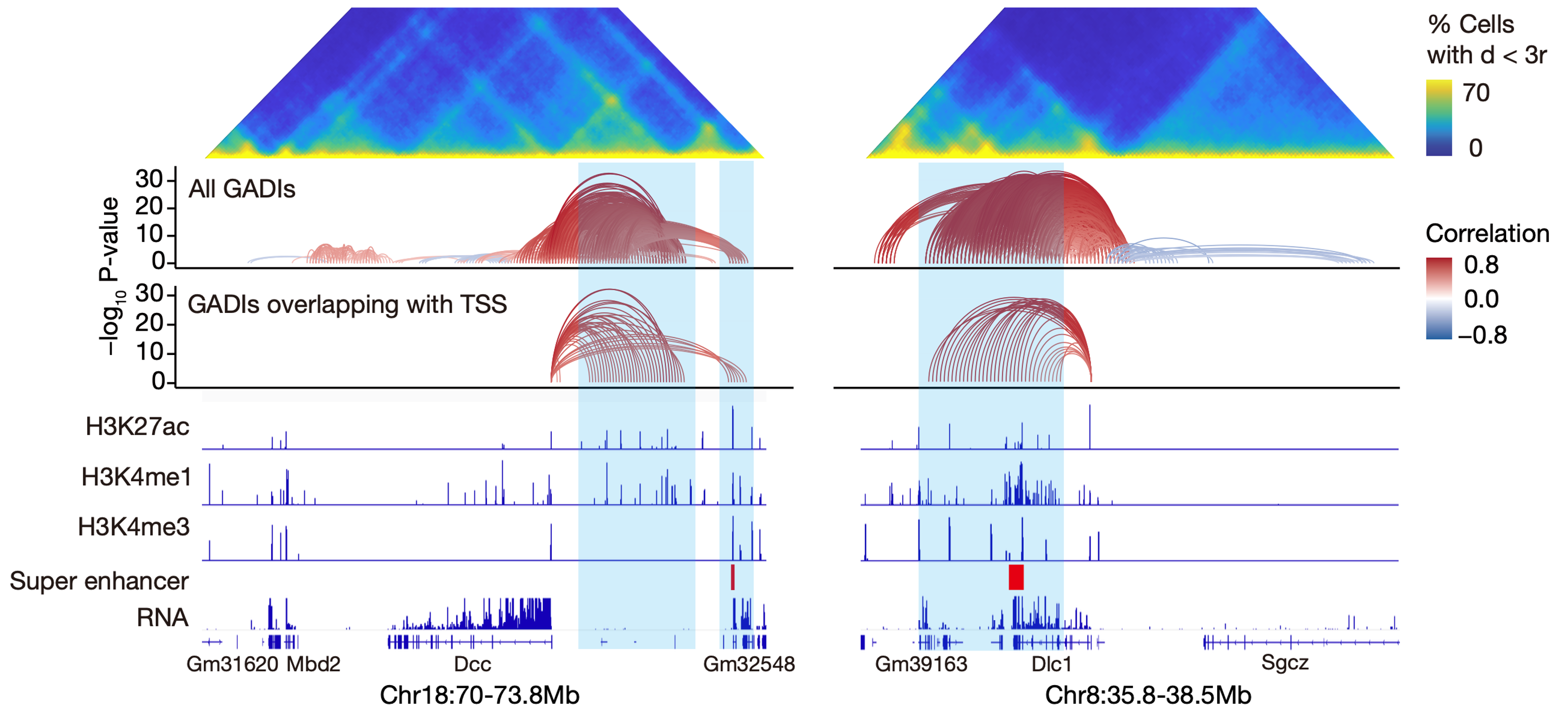
Figure 3 Example GADIs linking to Dcc(left) or Dlc1(right)
3. Discovery of widespread chromatin structural rearrangements preceding gene transcription activation
After linking chromatin interactions to specific genes with GADIs, the study further compared the pseudotemporal dynamics of chromatin conformation with transcription at high resolution and identified over 40,000 chromatin structural changes preceding transcriptional changes. Through epigenetic analysis of these structural change sites in different lineages, it was found that these chromatin structural changes preceding transcription mainly fall into two categories.
First, at most genetic loci, chromatin rewiring before gene expression was mainly established between active chromatin, which was possibly driven by the activation of enhancers. Second, for genes located in a repressed chromatin environment, it was the relaxation of the condensed heterochromatin that occurred before transcription activation (Figure 4). Although understanding the detailed molecular mechanisms behind these processes will require further experiments, this work have provided strong evidence for the regulatory role of the 3D genome on gene activation.
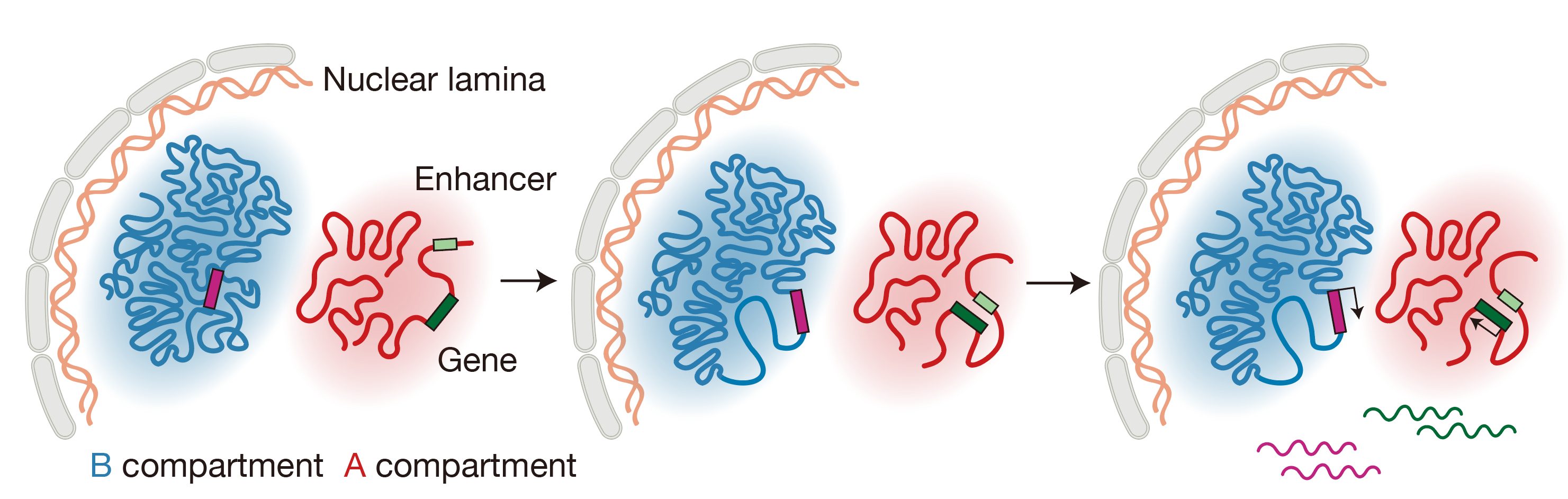
Figure 4 Schematics depicting chromatin reorganizations that occur before transcription activation
This work was supported by the Ministry of Science and Technology of China (grant STI2030 Major Projects 2021ZD0202502), the National Natural Science Foundation of China (grant 92049110), the Fundamental Research Funds for the Central Universities (Clinical Medicine Plus X Young Scholars Project, Peking University), and the Beijing Advanced Innovation Center for Genomics (ICG).
Link:https://www.science.org/doi/10.1126/science.adg3797
References:
1 Bas Tolhuis, R.-J. P., Erik Splinter, & Frank Grosveld, a. W. d. L. Looping and Interaction between Hypersensitive Sites in the Active beta-globin Locus. Molecular Cell (2002).
2 Deng, W. et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell 158, 849-860 (2014). https://doi.org:10.1016/j.cell.2014.05.050
3 Tang, Z. et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell 163, 1611-1627 (2015). https://doi.org:10.1016/j.cell.2015.11.024
4 Lupianez, D. G. et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012-1025 (2015). https://doi.org:10.1016/j.cell.2015.04.004
5 Flavahan, W. A. et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110-114 (2016). https://doi.org:10.1038/nature16490
6 Xu, Z. et al. Structural variants drive context-dependent oncogene activation in cancer. Nature 612, 564-572 (2022). https://doi.org:10.1038/s41586-022-05504-4
7 Nora, E. P. et al. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 169, 930-944 e922 (2017). https://doi.org:10.1016/j.cell.2017.05.004
8 Rao, S. S. P. et al. Cohesin Loss Eliminates All Loop Domains. Cell 171, 305-320 e324 (2017). https://doi.org:10.1016/j.cell.2017.09.026
9 Wutz, G. et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J 36, 3573-3599 (2017). https://doi.org:10.15252/embj.201798004
10 Ing-Simmons, E. et al. Independence of chromatin conformation and gene regulation during Drosophila dorsoventral patterning. Nat Genet 53, 487-499 (2021). https://doi.org:10.1038/s41588-021-00799-x
11 Espinola, S. M. et al. Cis-regulatory chromatin loops arise before TADs and gene activation, and are independent of cell fate during early Drosophila development. Nat Genet 53, 477-486 (2021). https://doi.org:10.1038/s41588-021-00816-z